More Than Just Taste and Smell: Examining the Complexity and Diversity of Terpene Profiles from Popular Cannabis Strains
Terpene profile analysis of terpene COAs
The title above was the running title of a poster I presented at the 2017 Cannabis Research Conference in CSU in Pueblo Colorado, the slides are available for download here.
Feel free to skip the PDF as we take a quick jaunt and describe my poster in more detail below.
Since we are story telling here I intend to walk you through each slide, but to keep things scientific this post will follow an academic format whereby the sections are divided into
- Abstract
- Methods
- Results
- Summary
- References
Abstract
Cannabis synthesizes a diversity of secondary metabolites. Selection and breeding has resulted in strains with enhanced cannabinoid potency and unique flavor profiles called chemotypes.
Volatile organic compounds known as terpenes are responsible for the taste and smell of cannabis are becoming recognized as medicinal molecules in their own right.
Our goal here is to create a botanical classification system to delineate chemotype groups and subgroups in popular strains using terpene profiles gathered from online sources.
When used in conjunction with genetic data this approach may be used to disambiguate or place the lineage of unknown strains and guide future breeding projects for the development of unique chemotypes with specific medicinal outcomes in mind.
Methods
-
Download Certificates Of Analysis (COAs) containing GC/MS quantiative terpene data from online sources (True Terpenes and archive.analytical360.com)
-
Extract text from PDF files on the command line using a Bash script
-
Clean raw data from text extraction using command line tools in Bash
-
Graph and Visualize dataset in the R programming language
Results
The "True Terpene" Dataset reveals a popular curation of strain terpene profiles
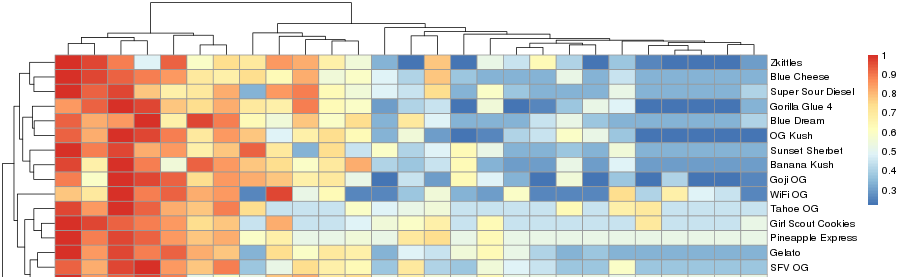
Clustering of the cannabis terpene "True Terpene" (COAs) dataset groups related clades and reveals similar chemotypes for several strains (Figure 1, below).
The above figure and quantitative terpene information for each strain can be visualized as a histogram by clicking here
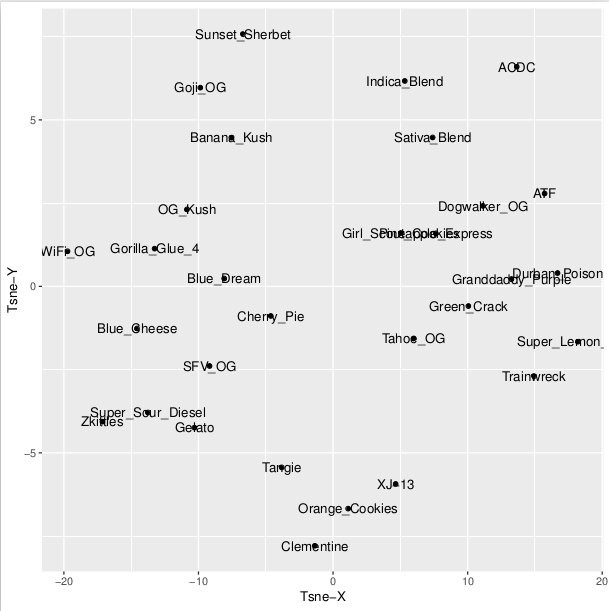
The True Terpene dataset can also be visualized using Principle Component Analysis (PCA) clustering similar profiles together and dissimilar profiles farther apart based on the dis-similarity (Figure 2, below). The label plotting issues will be fixed shortly.
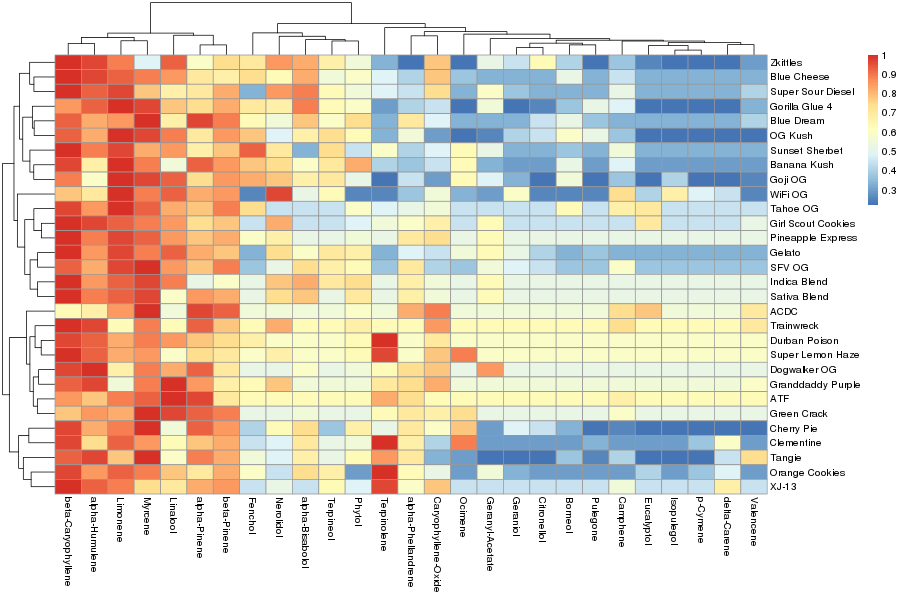
The terpene profiles of sour diesel and cheese strains share a distinct common chemotype (Figure 1 and 2, above and Figure 3 below).
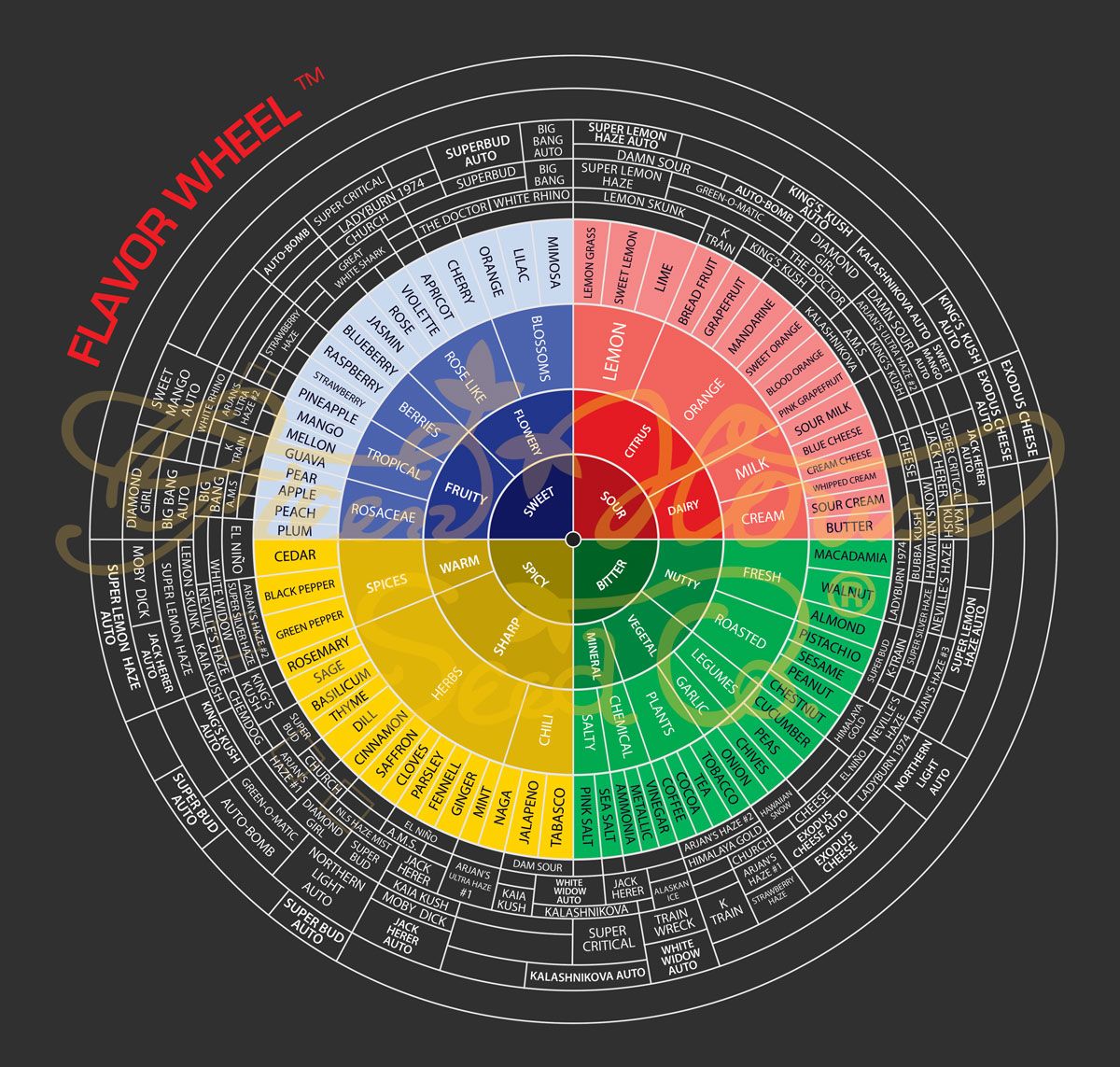
If you have not already seen the wonderful terpene wheel from Greenhouse Seed Company in the Netharlands you may want to take a moment to digest the following graphic below. Note that our fellow cannabis brethren have intuitively created a "Sour" category with a sub category called "Dairy" where we can find "Cheese" strains (top right quadrant of the image).
Sounds delicious doesn't it? Let's take a second and think about what I am saying and reference to your experience consuming cannabis. If you have consumed any Sour Diesel flower or product you know well that it has as Sour and Gas after taste that hmmm does taste like eaching a strong piece of cheese.
The Analytical 360 Dataset demonstrates intricate differences in terpene profiles within a strain name.
At this point you may or may not be thinking what are the "True Terpene" profiles of the strains tested by analytical 360. Well, let's take a look and see.
Let's start with my favorite strain Sour Diesel, see below.
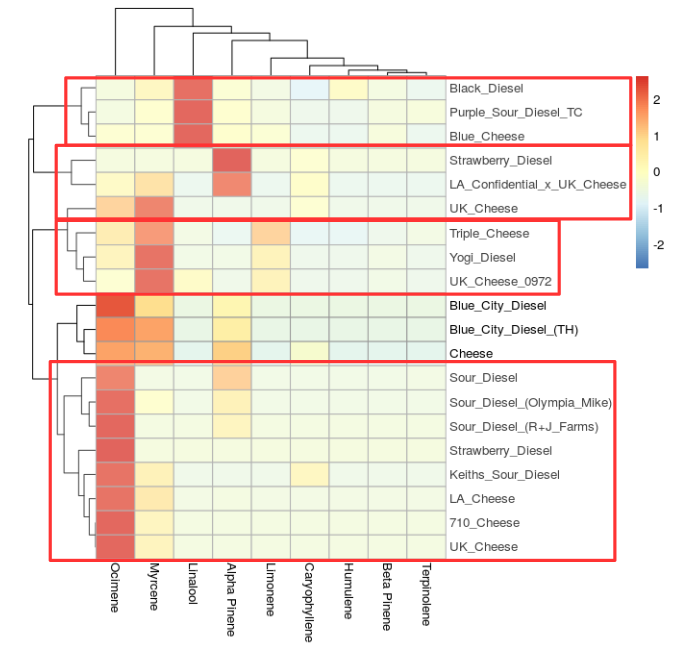
Recall, in Figure 1. the "cheese" and "diesel" strains clustered together. For this reason I have combined the two strains for comparison in Figure 3 (above), and in this more detailed report of all strains called "cheese" and "diesel".
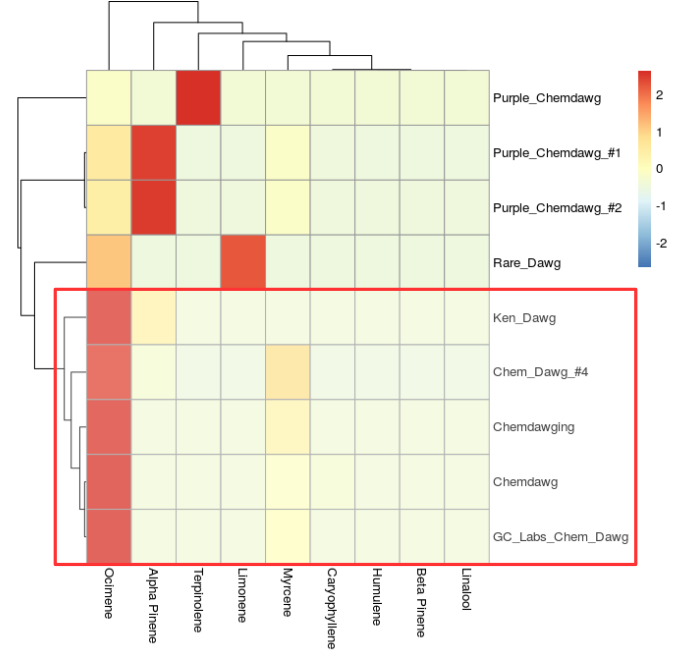
The popular strain Chemdawg has a distinct ocimene rich chemotype (Figure 4, below). Note "Purple chemdawg" has very little ocimene and "Rare dawg" has the highest limonene of the chemdawg samples. The customer who submitted "Rare Dawg" likely knew there was a subtle difference in the chemotype hence the name.
We could be even more fortunate if that same customer called it "Uplifting Dawg" or "Sativa Dawg", but no such luck here. In short, I feel that "Rare Dawg" would not be called "Dull Dawg" due to the high amount of limonene and low myrcene compared to the high alpha-pinene of "Purple chemdawg 1 and 2". These three strains may likely all lead to an alert/uplifting or "Sativa" high. Whether you believe in "Indica" or "Sativa" is another debate for another blog article, but I consider the term "Indica" useful to delineate sedative strains that produce "couch-lock" or put you "in-da couch". Whereas "Sativa" strains, here in this context as I have said above are the antithesis.
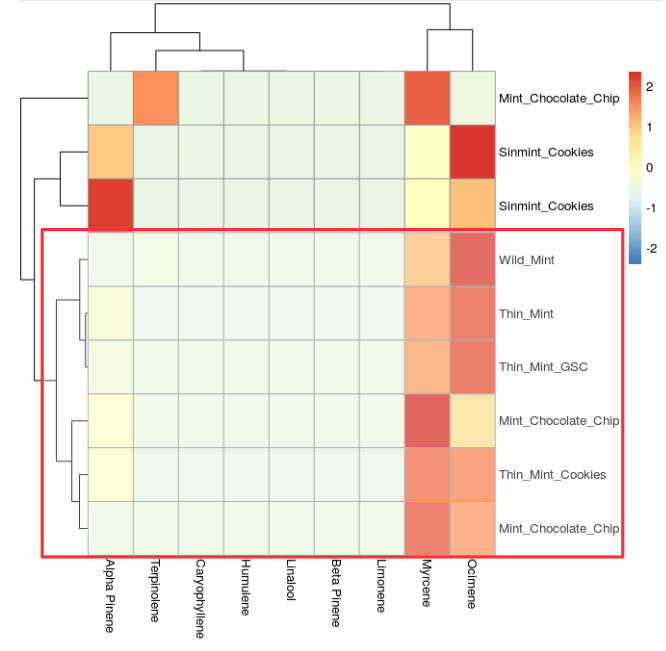
Thin Mint strains also have subtle variations in chemotypes (Figure 5, below). This may or may not have been inherited from the parental Girl Scout Cookies (GSC) strain since we see a Thin Mint GSC and Thin Mint have a very similar chemotype. A deep dive into the public geneology charts on Medicinal Genomics' strain database called Kannapedia could add more light to this trend we are observing.
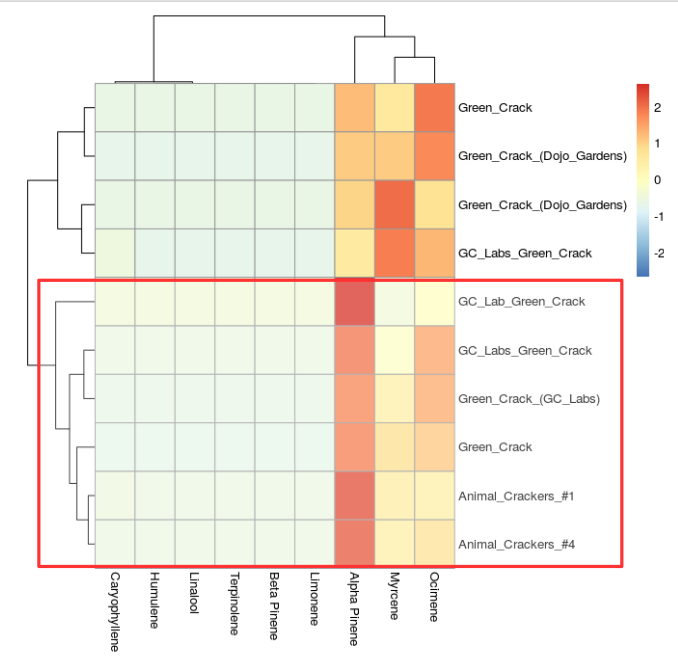
The "Green crack" strain, known by many as a "Super sativa" due to imparting a strong sense of enhanced alertness, wakefulness. and productivity after consumption appears to have a myrcene rich sub chemotype and a one that is more subdued. A cannabis sommelier may postulate that the myrcene depleted chemotype of "Green crack" would be more alert with less sedative effect associated directly with myrcene.
Notice the strain "GC Lab Green Crack" in the center is very low in myrcene (Figure 6, below). I would postulate this strain to be the most alert of the strains compared in this figure.
Conclusion
Computational classification of cannabis flower terpene data reveals strain specific chemotypes which can further be categorized into sub chemotypes. Genetic sequencing may be used to build a stronger hypothesis regarding the origin of strain specific chemotypes and sub chemotypes. Although, strains with similar chemotypes or sub chemotypes may have occured through a process of selection and convergent evolution rather than being closely related.
References
-
Fischedick, 2017. Identification of Terpenoid Chemotypes Among High (-)-trans-Detla-9-Tetrahydrocannabinol-Producing Cannabis sativa L. Cultivars.
-
Hazekamp et al., 2016. Cannabis: From Cultivar to Chemovar II—A Metabolomics Approach to Cannabis Classiffication
-
Henry, 2017. Cannabis chemovar classiffication: terpenes hyper-classes and targeted genetic markers for accurate discrimination of flavours and effects
-
Russo, 2010. Taming THC: potential cannabis synergy and phytocannabinoid-terpenoid entourage effects